In the previous post in this series, I showed how to use Azure OpenAI embedding models with Genkit Go and PostgreSQL’s pgvector extension using a Genkit Go sample. The required changes were minimal—registering the Azure OpenAI plugin and providing a Genkit embedder for the text-embedding-3-small model. The data access code for PostgreSQL remained completely unchanged.
But PostgreSQL (specifically Azure Database for PostgreSQL) isn’t the only managed database with vector search capabilities that Azure provides. In fact, every first-party managed database service in Azure (both relational and non-relational) offers some form of vector search capability.
So let’s look at how to build the same application using Azure SQL and its native vector support instead of PostgreSQL and pgvector.
Retrievers in Genkit Go#
Let’s review the concept of retrievers in Genkit and how it applies in the PostgreSQL sample.
A retriever is a Genkit plugin or a plain Go function that returns data from a vector store. Genkit offers several built-in retriever plugins for vector databases like its local dev store, Pinecone, and Weaviate.
The PostgreSQL sample doesn’t use a plugin. Instead, it uses an ad-hoc retriever function. A retriever function lets you write only the code needed to fetch your documents for your application. This code is likely less reusable than a proper plugin, but you don’t need to worry about building a reusable component at the start.
Here’s the code that sets up Genkit with the retriever function in the pgvector sample:
retOpts := &ai.RetrieverOptions{
ConfigSchema: nil,
Label: "pgVector",
Supports: &ai.RetrieverSupports{
Media: false,
},
}
retriever := defineRetriever(g, db, embedder, retOpts)
type input struct {
Question string
Show string
}
The code first creates RetrieverOptions. A retriever plugin must provide a way for the caller to limit the number of search results, set filters, or configure the vector distance calculation method, and that’s the purpose of RetrieverOptions. In the pgvector sample, this is just a technicality. All search details are hard-coded in the retriever function, so the sample creates a stub configuration.
Next, defineRetriever() is called to create and register the retriever function with Genkit. We’ll look at that in detail shortly.
With the retriever function registered, the code then defines the input struct. This type defines the input for the Genkit flow askQuestion, which is defined next.
genkit.DefineFlow(g, "askQuestion", func(ctx context.Context, in input) (string, error) {
res, err := genkit.Retrieve(ctx, g,
ai.WithRetriever(retriever),
ai.WithConfig(in.Show),
ai.WithTextDocs(in.Question))
if err != nil {
return "", err
}
for _, doc := range res.Documents {
fmt.Printf("%+v %q\n", doc.Metadata, doc.Content[0].Text)
}
// Use documents in RAG prompts.
return "", nil
})
The flow starts by calling genkit.Retrieve(), passing the Question field from our input through the ai.WithTextDocs() function, and the Show field as a ConfigOption through ai.WithConfig(). What’s the difference? Question is our search term and the input for the embedder. Show is used as a filter in our database query.
This logic remains the same regardless of which database we’re using in our retriever function, so we can leave this code almost unchanged. We will update the RetrieveOptions.Label later, but this a mere cosmetic change—the label isn’t used anywhere else in the code.
Show and adjust the query in the retriever function depending on which filters must be applied.The remaining code loops over the retrieval result and writes each document to the console—it doesn’t even return any result. That’s sufficient for a sample application you can run with the Genkit CLI, but in a retrieval-augmented generation (RAG) application, you would now craft the prompt including the relevant documents returned by genkit.Retrieve() and generate output using an LLM like GPT-5.
Now, let’s have a look at the retriever function defined in defineRetriever():
f := func(ctx context.Context, req *ai.RetrieverRequest) (*ai.RetrieverResponse, error) {
eres, err := genkit.Embed(ctx, g,
ai.WithEmbedder(embedder),
ai.WithDocs(req.Query))
if err != nil {
return nil, err
}
...
}
At runtime, Genkit provides the query information, i.e., the input provided through ai.WithTextDocs() in the call to genkit.Retrieve() as the Query field in the ai.RetrieverRequest. So when we invoke the flow as
genkit flow:run askQuestion '{"Show": "Best Friends", "Question": "Who does Alice love?"}'
the value of our input’s Question field becomes the value of RetrieverRequest.Query.
Next, the code calculates the vector embedding for Query using genkit.Embed(). Using the pgvector extension’s types and operators, the code queries the PostgreSQL database for the two most semantically similar records that match the show provided as a filter:
rows, err := db.QueryContext(ctx, `
SELECT episode_id, season_number, chunk as content
FROM embeddings
WHERE show_id = $1
ORDER BY embedding <#> $2
LIMIT 2`,
req.Options, pgv.NewVector(eres.Embeddings[0].Embedding))
if err != nil {
return nil, err
}
defer rows.Close()
From a Go perspective, this is standard data access code using the database/sql package and the PostgreSQL driver. The only specialty here is the use of the pgv package to provide a Go native representation of pgvector’s vector data type, and the use of the <#> operator to calculate the dot product of the input vector and a stored vector.
The query uses two positional parameters:
req.Optionsfor theWHEREclause. Remember that ourinputstruct for the Genkit flow also has theShowfield as a filter? This filter is provided through theOptionsfield.- The vector embedding calculated for the search term to order the query results by the dot product.
The query’s row set is then translated to an ai.RetrieverResponse which holds a slice of ai.Document pointers:
res := &ai.RetrieverResponse{}
for rows.Next() {
var eid, sn int
var content string
if err := rows.Scan(&eid, &sn, &content); err != nil {
return nil, err
}
meta := map[string]any{
"episode_id": eid,
"season_number": sn,
}
doc := &ai.Document{
Content: []*ai.Part{ai.NewTextPart(content)},
Metadata: meta,
}
res.Documents = append(res.Documents, doc)
}
if err := rows.Err(); err != nil {
return nil, err
}
return res, nil
The code scans every row in the row set, reads the fields we are interested in, and then creates a Document with the result’s content, storing episode_id and season_number as document metadata.
And these are the only PostgreSQL-specific parts of the retriever function: the database query and the mapping of the row set to a RetrieverResponse. We must update the query to use Azure SQL’s T-SQL syntax and vector functions, but the mapping code can remain unchanged as long as we use the same column names for our data model in Azure SQL.
Now that we’ve understood how Genkit interacts with the underlying database, let’s update the code so it works with Azure SQL instead.
Replacing PostgreSQL with Azure SQL#
Updating the Database Driver#
First, let’s switch to the correct database driver and create a SQL script to create our data model and sample data. To switch to Azure SQL, we can add the module with Microsoft’s official Go driver.
go get -u github.com/microsoft/go-mssqldb
Switching the driver means changing the driver name in the call to sql.Open():
db, err := sql.Open(azuread.DriverName, *connString)
if err != nil {
return err
}
defer db.Close()
Updating the SQL Script#
Next, we update the script that creates the table and inserts some sample data. Unlike PostgreSQL, Azure SQL’s vector features are built-in, so we don’t need to worry about extensions.
CREATE TABLE embeddings (
show_id NVARCHAR(255) NOT NULL,
season_number INT NOT NULL,
episode_id INT NOT NULL,
chunk NVARCHAR(MAX),
embedding VECTOR(1536),
PRIMARY KEY (show_id, season_number, episode_id)
);
INSERT INTO embeddings (show_id, season_number, episode_id, chunk) VALUES
('La Vie', 1, 1, 'Natasha confesses her love for Pierre.'),
('La Vie', 1, 2, 'Pierre and Natasha become engaged.'),
('La Vie', 1, 3, 'Margot and Henri divorce.'),
('Best Friends', 1, 1, 'Alice confesses her love for Oscar.'),
('Best Friends', 1, 2, 'Oscar and Alice become engaged.'),
('Best Friends', 1, 3, 'Bob and Pat divorce.')
;
Updating the RetrieverOptions#
Now, let’s update the label of the RetrieverOptions—this is a cosmetic change.
retOpts := &ai.RetrieverOptions{
ConfigSchema: nil,
Label: "azureSQL",
Supports: &ai.RetrieverSupports{
Media: false,
},
}
...
Updating the Retriever Function#
Next up are the interesting bits: We must update the retriever function to query Azure SQL.
f := func(ctx context.Context, req *ai.RetrieverRequest) (*ai.RetrieverResponse, error) {
eres, err := genkit.Embed(ctx, g,
ai.WithEmbedder(embedder),
ai.WithDocs(req.Query))
if err != nil {
return nil, err
}
vector, err := json.Marshal(eres.Embeddings[0].Embedding)
if err != nil {
return nil, err
}
rows, err := db.QueryContext(ctx, `
SELECT TOP(2) episode_id, season_number, chunk as content
FROM embeddings
WHERE show_id = @show_id
ORDER BY VECTOR_DISTANCE('dot', embedding, CAST(@embedding AS VECTOR(1536)))
`,
sql.Named("show_id", req.Options),
sql.Named("embedding", string(vector)))
if err != nil {
return nil, err
}
defer rows.Close()
...
}
The first notable change here is that we create a JSON representation of the embedding. Recall that the pgvector version of the code uses a pgv.Vector instance. This type wraps the underlying []float32 that stores the vector’s dimensions and is used by Go’s PostgreSQL driver to map it to its native vector(1536) type. At the time of writing, the Microsoft SQL Go driver doesn’t support a Go native representation of vectors. Instead, it expects vectors to be passed as NVARCHAR(MAX) with a JSON representation of the vector. Hence, we marshal the vector to JSON.
The next change is the query itself. Besides the required syntax change from LIMIT 2 to TOP(2), I chose to use named parameters instead of positional parameters, but that’s merely a personal preference.
Unlike pgvector, which defines operators for vector distance calculations, Azure SQL provides the VECTOR_DISTANCE function. The first parameter expected by this function is the distance metric. We’re using dot to calculate the dot product, like the PostgreSQL version did using the <#> operator. We also must type cast our embedding parameter to a VECTOR(1536).
As mentioned before, the code to map the query’s row set to a RetrieverResponse does not need to be changed, since we’re using the same column names.
Updating the Indexing Feature#
The pgvector sample also contains a feature to index the test data in the database:
func indexExistingRows(ctx context.Context, g *genkit.Genkit, db *sql.DB, embedder ai.Embedder) error {
rows, err := db.QueryContext(ctx, `SELECT show_id, season_number, episode_id, chunk FROM embeddings`)
if err != nil {
return err
}
defer rows.Close()
var docs []*ai.Document
for rows.Next() {
var sid, chunk string
var sn, eid int
if err := rows.Scan(&sid, &sn, &eid, &chunk); err != nil {
return err
}
docs = append(docs, &ai.Document{
Content: []*ai.Part{ai.NewTextPart(chunk)},
Metadata: map[string]any{
"show_id": sid,
"season_number": sn,
"episode_id": eid,
},
})
}
if err := rows.Err(); err != nil {
return err
}
return Index(ctx, g, db, embedder, docs)
}
There is nothing new here. The code iterates through the data set, creates an ai.Document per row using show_id, season_number, and episode_id as metadata, and invokes the Index() function. The indexExistingRows() function works as is for Azure SQL, since the query doesn’t use any PostgreSQL-specific features or syntax.
The Index() function, on the other hand, has PostgreSQL-specific code. It invokes the embedder for every Document passed by indexExistingRows, extracts its metadata (show_id, season_number, and episode_id), and then executes an UPDATE statement with the matching parameters for the WHERE clause.
func Index(ctx context.Context, g *genkit.Genkit, db *sql.DB, embedder ai.Embedder, docs []*ai.Document) error {
// The indexer assumes that each Document has a single part, to be embedded, and metadata fields
// for the table primary key: show_id, season_number, episode_id.
const query = `
UPDATE embeddings
SET embedding = $4
WHERE show_id = $1 AND season_number = $2 AND episode_id = $3
`
res, err := genkit.Embed(ctx, g,
ai.WithEmbedder(embedder),
ai.WithDocs(docs...))
if err != nil {
return err
}
// You may want to use your database's batch functionality to insert the embeddings
// more efficiently.
for i, emb := range res.Embeddings {
doc := docs[i]
args := make([]any, 4)
for j, k := range []string{"show_id", "season_number", "episode_id"} {
if a, ok := doc.Metadata[k]; ok {
args[j] = a
} else {
return fmt.Errorf("doc[%d]: missing metadata key %q", i, k)
}
}
args[3] = pgv.NewVector(emb.Embedding)
if _, err := db.ExecContext(ctx, query, args...); err != nil {
return err
}
}
return nil
}
As with the retriever method, I prefer to use named parameters for the query, so let’s update the query first:
const query = `
UPDATE embeddings
SET embedding = @embedding
WHERE show_id = @show_id AND season_number = @season_number AND episode_id = @episode_id
`
Because we use named parameters, we have to create them when extracting the metadata, not just store the parameter value:
for j, k := range []string{"show_id", "season_number", "episode_id"} {
if a, ok := doc.Metadata[k]; ok {
args[j] = sql.Named(k, a)
} else {
return fmt.Errorf("doc[%d]: missing metadata key %q", i, k)
}
}
And last but not least, we have to apply the JSON string workaround for the vector.
vector, err := json.Marshal(emb.Embedding)
if err != nil {
return err
}
args[3] = sql.Named("embedding", string(vector))
We’re done! These are all the changes required to switch from PostgreSQL and pgvector to Azure SQL. Now all you need is an Azure SQL database and to apply your database script.
Running the Sample#
Assuming you already have a running Azure SQL database, run the database script with your preferred database client to create the data model and seed the sample data. The GitHub repository for this blog post contains a script to bootstrap an Azure SQL database if you want to deploy a demo instance.
Open a terminal window and run the application. Make sure to pass your database’s connection string using the -dbconn flag.
export AZ_OPENAI_BASE_URL='https://<your-resource>.openai.azure.com/openai/v1/'
export AZ_OPENAI_API_KEY='<your-azure-openai-api-key>'
export GENKIT_ENV='dev'
# The init flag triggers the embedding generation
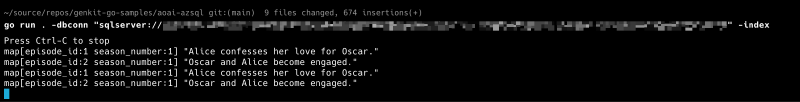
go run . -dbconn "sqlserver://<username>:<password>@<servername>.database.windows.net?database=<database-name>" -index
From another terminal, trigger the vector search flow:
genkit flow:run askQuestion '{"Show": "La Vie", "Question": "Who gets divorced?"}'
genkit flow:run askQuestion '{"Show": "Best Friends", "Question": "Who does Alice love?"}'
The vector search results aren’t returned as flow output—the sample logs the raw Go structs to the console where your application runs, showing the retrieved documents.
Summary#
This post explained how to use Azure OpenAI embeddings and Azure SQL’s native vector search with Genkit Go. It covered the retriever concept, the required code, and practical steps for setup and execution. The article demonstrated how Genkit can flexibly integrate LLMs with enterprise data sources using Azure SQL.